Working Example
working_example.Rmd
library(MinervaTime)See the reference section for notes on individual functions.
This working example contains 10 trials. On each trial an A event is presented for 25 time steps, followed by a B event for 25 time steps. The onset of the B event is randomly determined by a normal distribution, centered on the 50th time step, with a standard deviation of 10 timesteps.
Create trials
#### BUILD TRAINING SCENARIO
# e.g., a matrix of environment vectors
# representing events unfolding over time within a trial
# with the capability to train multiple trials
trial_timesteps <- 200 # define number of time steps per trial
# create a timeline of trials, listing temporal event properties per trial
timeline <- make_event_timeline(
num_trials = 10,
events = c('A', 'B', 'C'),
onsets = list(1,round(rnorm(10,50,10)),1),
durations = list(25,25,199)
)
# convert timeline to environment vectors
temporal_vectors <- make_temporal_vectors(trial_timesteps, overlap = 2)
event_vectors <- make_event_vectors(LETTERS[1:10],10)
timeline_vectors <- timeline_to_vector(timeline,
event_vectors,
temporal_vectors,200)Run model
## define basic model parameters
library(RsemanticLibrarian) # for cosine_x_to_m
# initialize memory with 5 noise vectors
environment_matrix <- timeline_vectors[[1]]
noise <- matrix(runif(5*dim(environment_matrix)[2],-1,1)*.05,
nrow = 5,
ncol = dim(environment_matrix)[2])
#memory <- matrix(0,
# nrow=dim(environment_matrix)[1],
# ncol=dim(environment_matrix)[2])
memory <- noise
tau <- 3
## Run the model for each trial
model_results <- list()
for(t in 1:length(timeline_vectors)) {
environment_matrix <- timeline_vectors[[t]]
save_echo <- matrix(0,
nrow=dim(environment_matrix)[1],
ncol=dim(environment_matrix)[2])
# intra-trial encoding and retrieval
for (i in 1:dim(environment_matrix)[1]){
activations <- c(cosine_x_to_m(environment_matrix[i,],
memory)^tau)
echo <- colSums(memory*activations)
save_echo[i,] <- echo
memory <- rbind(memory,environment_matrix[i,])
}
A_expectation <- rowMeans(save_echo[,1:10])
#plot(A_expectation)
B_expectation <- rowMeans(save_echo[,11:20])
#plot(B_expectation)
model_results[[t]] <- list(A_expectation = A_expectation,
B_expectation = B_expectation,
save_echo = save_echo)
}plot model results
Shows the model’s expectation for the B event across a trial window, after the 1st, 5th, and 10th trial.
plot(model_results[[1]]$B_expectation)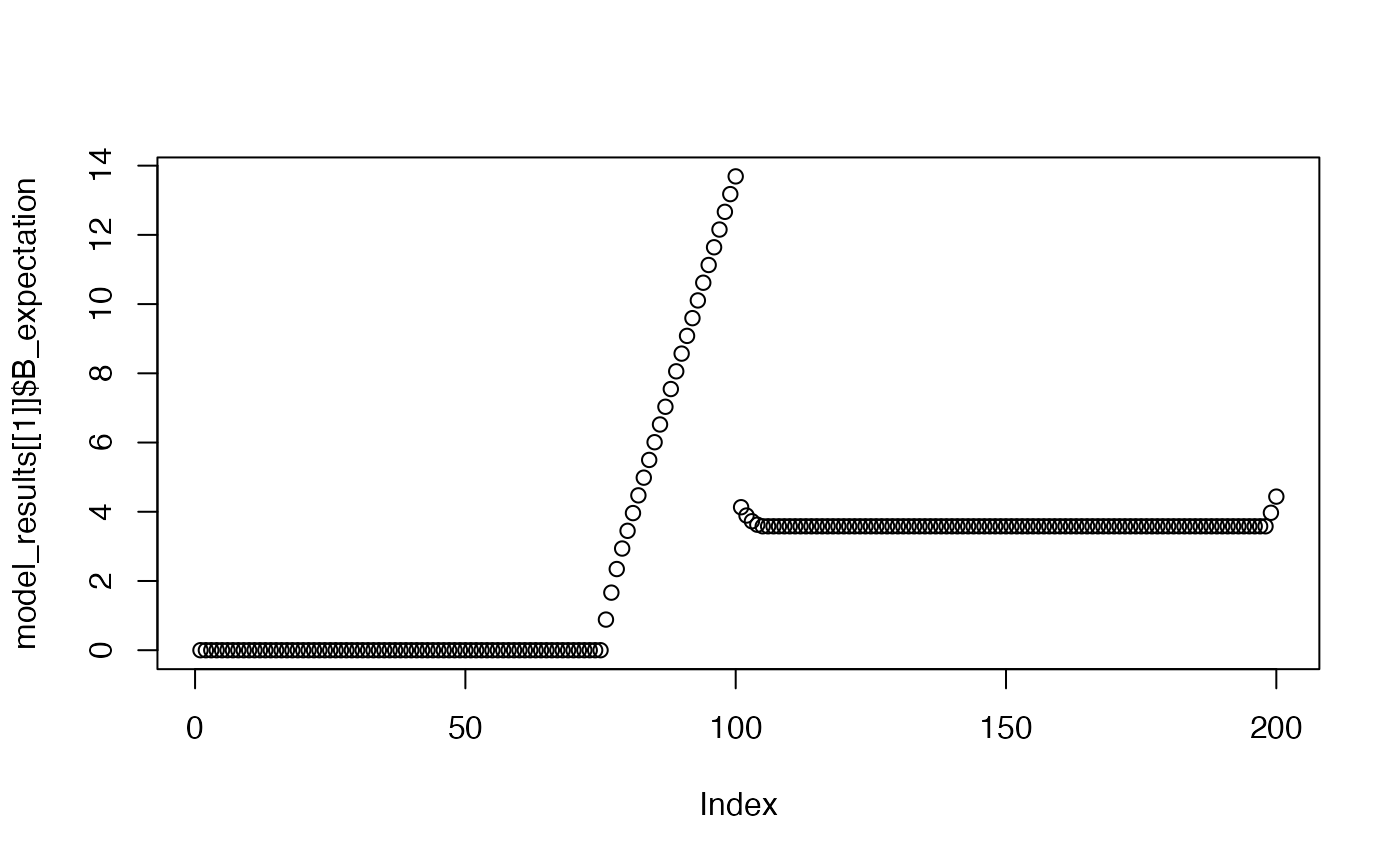
plot(model_results[[5]]$B_expectation)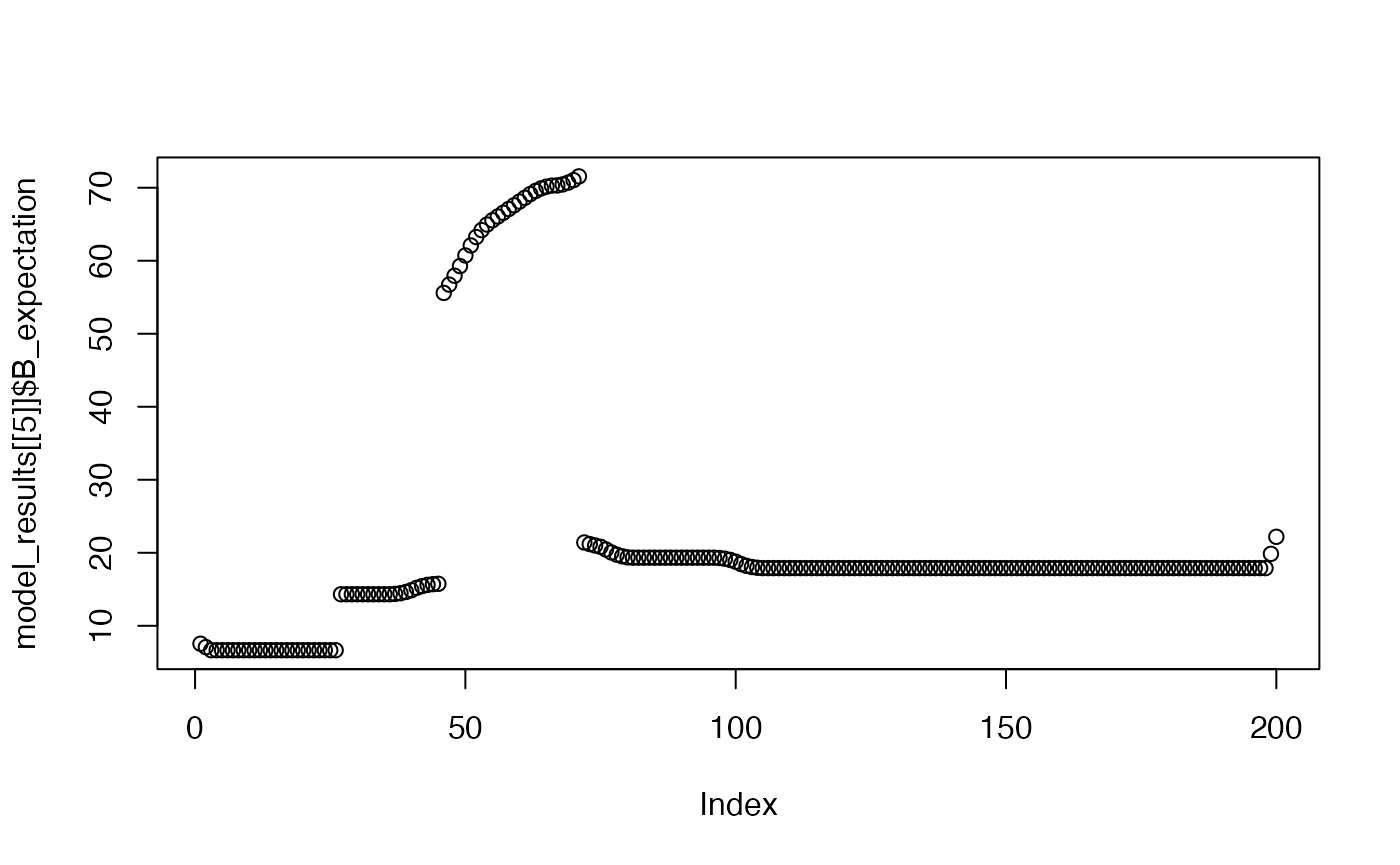
plot(model_results[[10]]$B_expectation)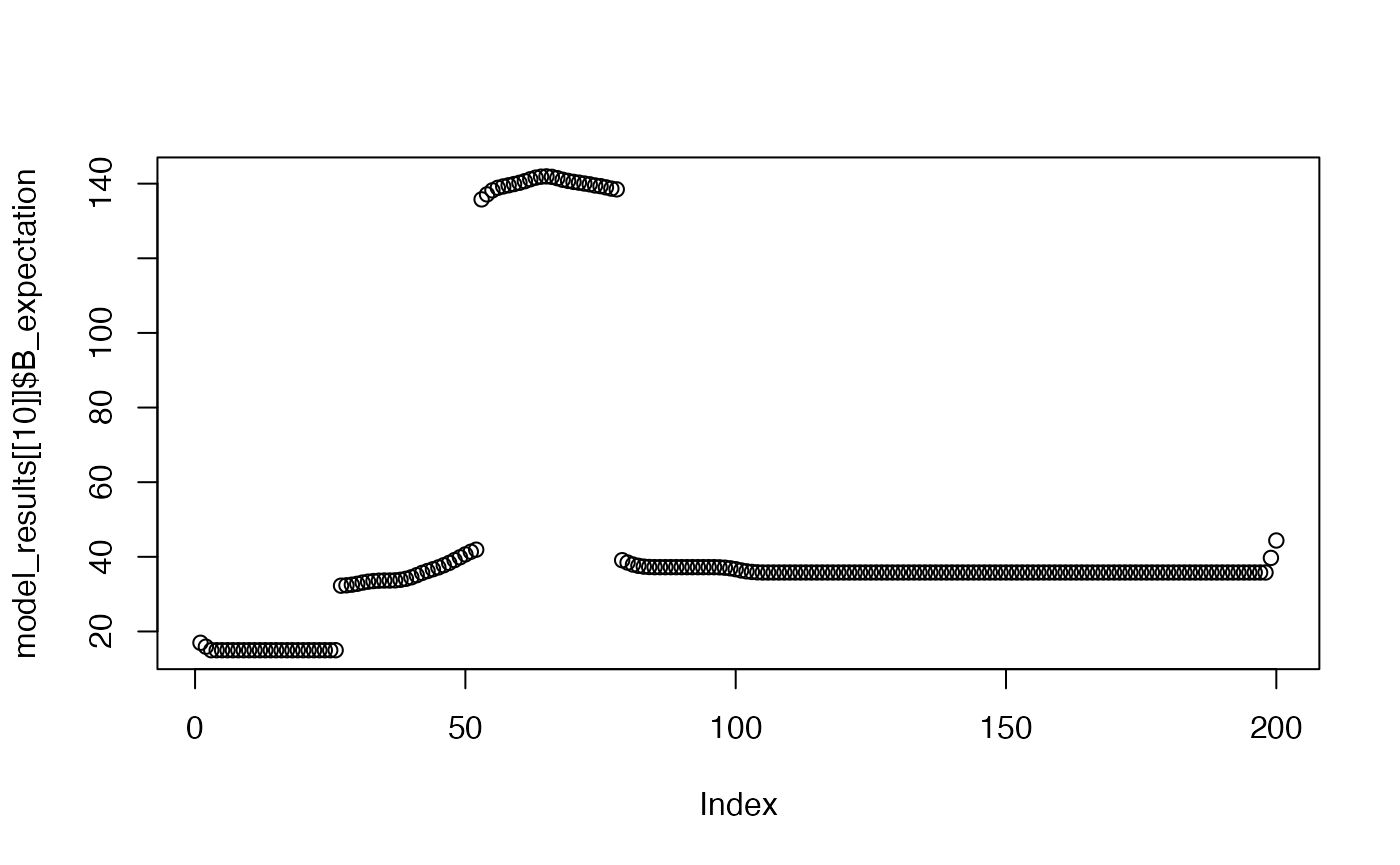
Iterative retrieval
tau <- 3
get_echo <- function(probe,memory,tau){
activations <- cosine_x_to_m(probe,memory)
weighted_memory <- memory*c(activations)^tau
echo <- colSums(weighted_memory)
norm_echo <- echo/max(abs(echo))
return(norm_echo)
}
probe <- c(rep(c(1,0,1),each=10), temporal_vectors[1,])
echo <- get_echo(probe,memory, 3)
plot(echo)
echo_2 <- get_echo(echo, memory, 3)
plot(echo_2)
echo_3 <- get_echo(echo_2, memory, 3)
plot(echo_3)
echo_4 <- get_echo(echo_3, memory, 3)
plot(echo_4)
echo_5 <- get_echo(echo_4, memory, 3)
plot(echo_5)
Try new stuff
Run model with discrepancy encoding
## define basic model parameters
library(RsemanticLibrarian) # for cosine_x_to_m
# initialize memory with 5 noise vectors
environment_matrix <- timeline_vectors[[1]]
noise <- matrix(runif(5*dim(environment_matrix)[2],-1,1)*.05,
nrow = 5,
ncol = dim(environment_matrix)[2])
#memory <- matrix(0,
# nrow=dim(environment_matrix)[1],
# ncol=dim(environment_matrix)[2])
memory <- noise
tau <- 3
## Run the model for each trial
model_results <- list()
for(t in 1:length(timeline_vectors)) {
environment_matrix <- timeline_vectors[[t]]
save_echo <- matrix(0,
nrow=dim(environment_matrix)[1],
ncol=dim(environment_matrix)[2])
# intra-trial encoding and retrieval
for (i in 1:dim(environment_matrix)[1]){
activations <- c(cosine_x_to_m(environment_matrix[i,],
memory)^tau)
echo <- colSums(memory*activations)
echo <- echo/max(abs(echo))
save_echo[i,] <- echo
memory <- rbind(memory,(echo-environment_matrix[i,]))
}
A_expectation <- rowMeans(save_echo[,1:10])
#plot(A_expectation)
B_expectation <- rowMeans(save_echo[,11:20])
#plot(B_expectation)
model_results[[t]] <- list(A_expectation = A_expectation,
B_expectation = B_expectation,
save_echo = save_echo)
}