#load libraries
library(tidyverse)
library(jsonlite)
library(xtable)
library(conflicted)
conflicts_prefer(dplyr::filter, .quiet = TRUE)
# directory
data_dir <- "data/jatos_results_data_20240305180329/"
# get subject data directories
sub_data_dirs <- list.files(data_dir)
all_data <- data.frame()
for(i in 1:length(sub_data_dirs)){
# get directory and data file name
component_dirs <- list.files(paste0(data_dir,"/",sub_data_dirs[i]))
get_file_name <- paste0(data_dir,"/",sub_data_dirs[i],"/",component_dirs[2],"/","data.txt")
# convert from json to data frame
read_file(get_file_name) %>%
# ... split it into lines ...
str_split('\n') %>% first() %>%
# ... filter empty rows ...
discard(function(x) x == '') %>%
# ... parse JSON into a data.frame
map_dfr(fromJSON, flatten=T) -> sub_data
# append to all_data
all_data <- rbind(all_data,sub_data)
}1a pilot analysis
This document contains example scripts for conducting an analysis of our pilot data. Each section contains basic steps along a data analysis pipeline. Roughly the steps are: import the data, clean or pre-process the data if necessary, check the data to make sure for issues (e.g., participants not complying with instructions, missing data, other issues), conduct planned analyses.
This is a quarto document, so it is possible to write plain text and to include code chunks for analysis. It is also possible to include inline R code, allowing us to print results from R objects into the document.
Another workflow aspect is that results will be stored in R objects. When this analysis file is “rendered”, all of code chunks will evaluate and producing R objects in a temporary environment. The last code chunk saves the environment (containing all R objects) to disk. This way, the results can be easily imported into the main manuscript file.
Import raw data
1a: Import data
Pre-processing
Use this stage to check data for irregularities.
row_counts <- all_data %>%
group_by(ID) %>%
count()proportion_response <- all_data %>%
filter(task == "judgment") %>%
mutate(response = as.numeric(response)) %>%
group_by(ID,bits) %>%
summarise(mean_choice = mean(response),.groups = "drop")
sub_perf <- proportion_response %>%
group_by(ID) %>%
summarise(m = mean(mean_choice),
s = sd(mean_choice),.groups = "drop")demographics
library(tidyr)
demographics <- all_data %>%
filter(trial_type == "survey-html-form") %>%
select(ID,response,counterbalance) %>%
unnest_wider(response) %>%
mutate(age = as.numeric(age))
age_demographics <- demographics %>%
summarize(mean_age = mean(age),
sd_age = sd(age),
min_age = min(age),
max_age = max(age))
factor_demographics <- apply(demographics[-1], 2, table)Print n per condition
# participants per condition
demographics %>%
group_by(counterbalance) %>%
summarize(condition_n = n(),.groups = "drop")# A tibble: 2 × 2
counterbalance condition_n
<chr> <int>
1 1a-feedback 24
2 1a-no-feedback 22pilot analysis
Individual Subject plots
# get only rows where a judgment was made
filtered_data <- all_data %>%
filter(task == "judgment")
# create data frame of proportions for each subject
sub_means <- filtered_data %>%
#convert to numeric
mutate(response = as.numeric(response)) %>%
group_by(counterbalance,ID,bits) %>%
summarize(mean_diff = mean(response),.groups = "drop")
ggplot(sub_means, aes(x = bits, y=mean_diff, color=ID))+
geom_point()+
geom_line()+
facet_wrap(~counterbalance)+
theme_classic() +
theme(legend.position="none")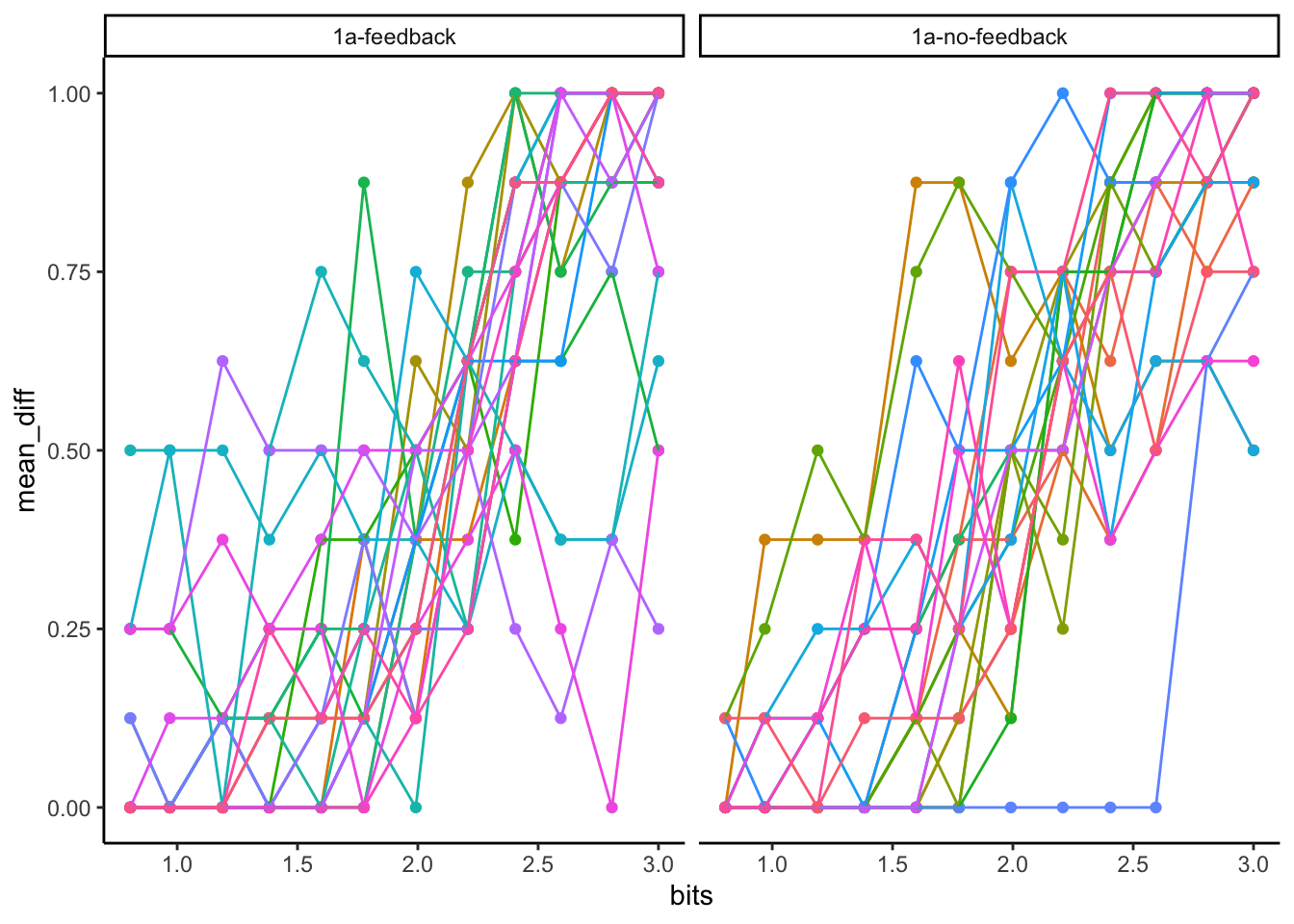
group means
Here we average over the individual subjects.
# average over subjects
group_means <- sub_means %>%
group_by(counterbalance,bits) %>%
summarize(mean_diff = mean(mean_diff),.groups = "drop")
#plot
ggplot(group_means, aes(x = bits, y=mean_diff))+
geom_point()+
facet_wrap(~counterbalance)+
theme_classic()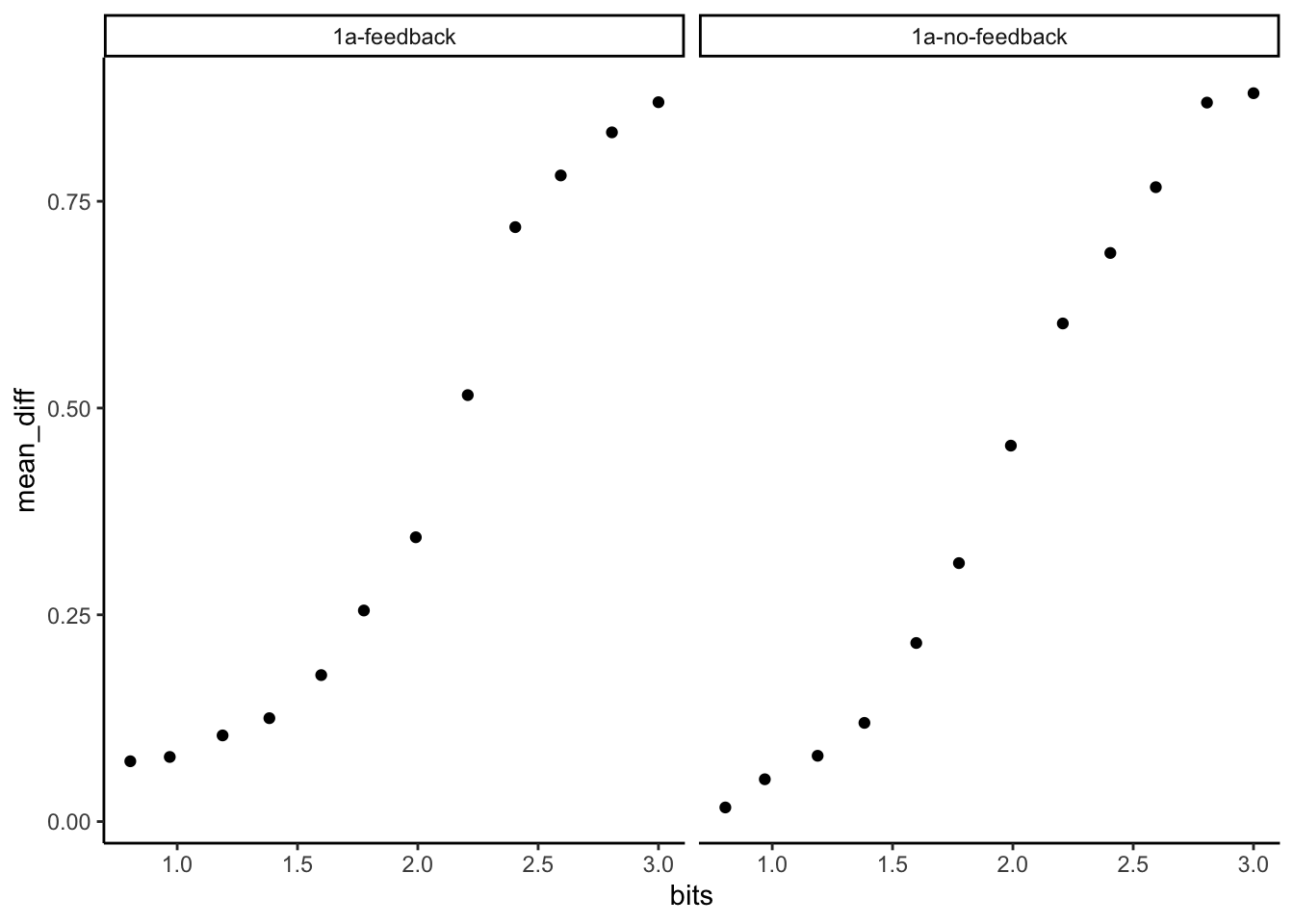
fitting a psychophysics function
library(quickpsy)
# include only rows where a response occurred
filtered_data_psy <- all_data %>%
filter(task == "judgment")
# format for quickpsy
sub_totals_psy <- filtered_data_psy %>%
mutate(response = as.numeric(response)) %>%
group_by(counterbalance,bits) %>%
summarize(sum_response = sum(response),
total_response = n(),
.group = "drop"
)
#fit curve
try_fit <- quickpsy(sub_totals_psy,
x = bits,
k = sum_response,
n = total_response,
guess = TRUE,
grouping = .(counterbalance))
# plot
plotcurves(try_fit)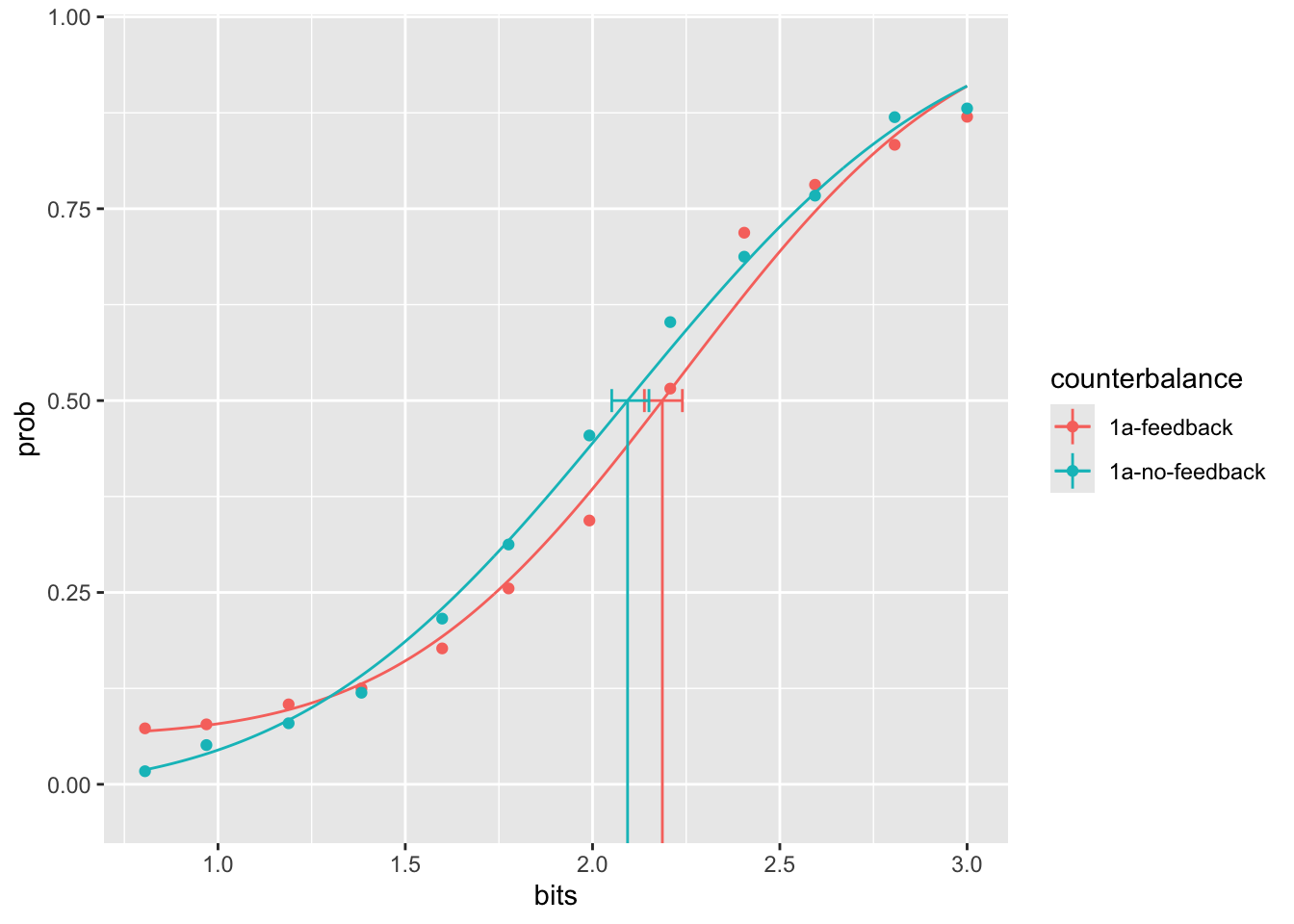
# print parameters
try_fit$par# A tibble: 6 × 5
# Groups: counterbalance [2]
counterbalance parn par parinf parsup
<chr> <chr> <dbl> <dbl> <dbl>
1 1a-feedback p1 2.24 2.17 2.30
2 1a-feedback p2 0.587 0.521 0.664
3 1a-feedback p3 0.0623 0.0288 0.0908
4 1a-no-feedback p1 2.08 2.02 2.15
5 1a-no-feedback p2 0.680 0.599 0.755
6 1a-no-feedback p3 -0.0116 -0.0430 0.0216save workspace
save.image("data/1a_analysis.RData")Using inline code to write results.
Inline code snippets allow one to write R results within a text segment. This is an R code snippet that prints the value of 1+1 = 2. We can use R code snippets to write results.
Psychophysical functions were fit to the data in each feedback condition using cumulative normal curves. The point of subject equality in the feedback condition was 2.19. Note this value was read in from an R variable and printed here. We will use this style of reporting in the main manuscript to make sure our results are reproducible.
Pilot cluster analysis
load("data/Sequence_matrix.RData")
# function to compute mean inter-item distance
mean_item_distance <- function(x){
x_to_char <- as.character(x)
all_unique <- unique(x_to_char)
counter <- rep(0,length(all_unique))
names(counter) <- all_unique
all_differences <- c()
for(i in 1:length(all_unique)){
differences <- diff( which(x_to_char %in% all_unique[i] == TRUE), diff = 1)
differences <- c(0,(differences-1))
all_differences <- c(all_differences,differences)
}
return(mean(all_differences))
}
clustering_scores <- apply(sequences_matrix,MARGIN = 1,mean_item_distance)
cluster_df <- data.frame(file_name = names(clustering_scores),
clustering = clustering_scores)
new_filtered <- filtered_data %>%
mutate(file_name = gsub("mp3s/","", stimulus)) %>%
mutate(file_name = gsub(".mp3",".mid",file_name)) %>%
mutate(response = as.numeric(response))
new_filtered <- left_join(new_filtered,cluster_df,'file_name')
ggplot(new_filtered, aes(x=clustering,y=response))+
geom_point()+
geom_smooth(method = "lm")+
facet_wrap(~bits, scales = "free")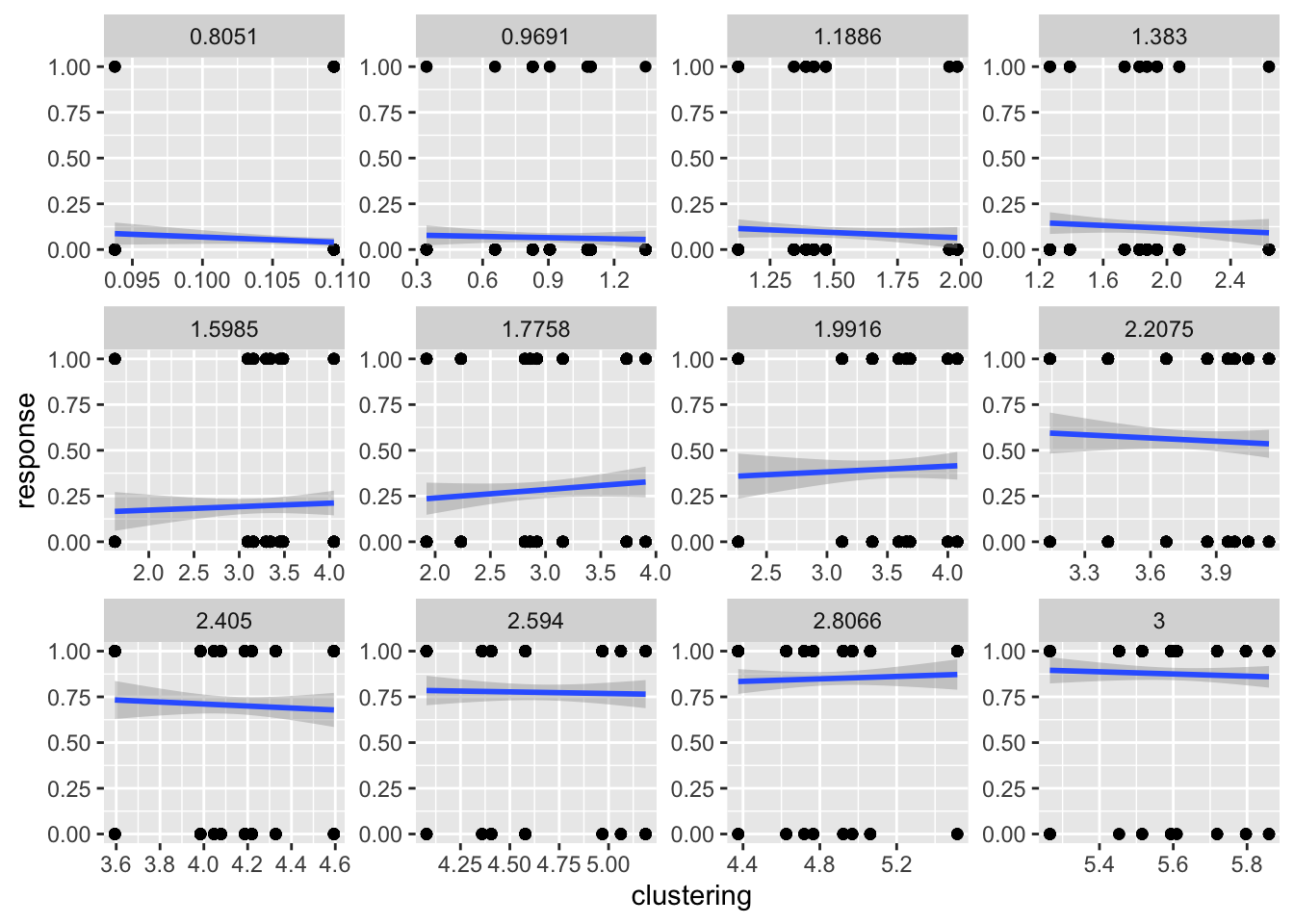
plot(new_filtered$bits,new_filtered$clustering)
sort(unique(new_filtered$bits)) [1] 0.8051 0.9691 1.1886 1.3830 1.5985 1.7758 1.9916 2.2075 2.4050 2.5940
[11] 2.8066 3.0000
comments