Show the code
library(tidyverse)
library(openai)
library(patchwork)
library(xtable)June 28, 2023
Started. Finished.
This is a GPT simulation of the SNARC effect, which is commonly observed in participants who have a mental number line with small to large numbers going from left to right.
A participant is presented with a digit from 1 to 9, and they judge whether the digit is odd or even with a right or left button press. People are faster to press the left button when the stimulus is a small number compared to a large number, and faster to press the right button when the stimulus is a large number compared to a small number.
Notes: 20 simulated subjects. 32 trials each. Stimuli were 1,2,3,4,6,7,8,9. Respond left for odd, right for even.
The prompt contains only basic information to complete the task.
Used gpt-3.5-turbo-16k, with max tokens 10000.
Problems: Still getting the occasional invalid JSON back, mostly due to the chatbot prefacing its response with a message before the JSON. This thread may be helpful https://community.openai.com/t/getting-response-data-as-a-fixed-consistent-json-response/28471/31?page=2.
# Use numbers 1 to 9.
snarc_items <- data.frame(stimulus = c(1,2,3,4,6,7,8,9),
instruction = "Respond left for odd, right for even numbers",
response = "?",
reaction_time = "?")
#set up variables to store data
all_sim_data <- tibble()
gpt_response_list <- list()
# request multiple subjects
# submit a query to open ai using the following prompt
# note: responses in JSON format are requested
for(i in 1:20){
print(i)
# construct trials data frame
snarc_trials <- snarc_items[rep(1:nrow(snarc_items),4),]
trials <- snarc_trials
trials <- trials[sample(1:nrow(trials)),]
trials <- trials %>%
mutate(trial = 1:nrow(trials)) %>%
relocate(instruction) %>%
relocate(trial)
# run the api call to openai
gpt_response <- create_chat_completion(
model = "gpt-3.5-turbo-16k",
max_tokens = 10000,
messages = list(
list(
"role" = "system",
"content" = "You are a simulated participant in a human cognition experiment. Complete the task as instructed and record your simulated responses in a JSON file. Do not include any explanations, only provide a RFC8259 compliant JSON response."),
list("role" = "assistant",
"content" = "OK, I am ready."),
list("role" = "user",
"content" = paste('You are a simulated participant in a human cognition experiment. Complete the task as instructed and record your simulated responses in JSON. Your task is to simulate human performance in a number judgment task. You will be given the task in the form a JSON object. The JSON object contains the task instruction and the stimulus on each trial. Your task is to follow the instruction and respond to the stimulus as quickly and accurately as a human participant would. The instructions are to identify the stimulus by responding left for odd numbers and right for even numbers. When you simulate data make sure it conforms to how humans would perform this task. The JSON object contains the symbol ? in locations where you will generate simulated responses. You will generate a simulated identification response, and a simulated reaction time for each trial. Put the simulated identification response and reaction time into a JSON array using this format: [{"trial": "trial number, integer", "instruction" = "the task instruction, string", "stimulus": "the stimulus, integer", "response": "the simulated identification response, string","reaction_time": "the simulated reaction time, milliseconds an integer"}].', "\n\n", jsonlite::toJSON(trials), collapse = "\n")
)
)
)
# save the output from openai
gpt_response_list[[i]] <- gpt_response
print(gpt_response$usage$total_tokens)
# validate the JSON
test_JSON <- jsonlite::validate(gpt_response$choices$message.content)
print(test_JSON)
# validation checks pass, write the simulated data to all_sim_data
if(test_JSON == TRUE){
sim_data <- jsonlite::fromJSON(gpt_response$choices$message.content)
if(sum(names(sim_data) == c("trial","instruction","stimulus","response","reaction_time")) == 5) {
sim_data <- sim_data %>%
mutate(sim_subject = i)
all_sim_data <- rbind(all_sim_data,sim_data)
}
}
}
# model responses are in JSON format
save.image("data/simulation_11.RData")The LLM occasionally returns invalid JSON. The simulation ran 20 times.
There were 17 out of 20 valid simulated subjects.
The model made too many mistakes to analyse RTs.
all_sim_data <- all_sim_data %>%
mutate(reaction_time = as.numeric(reaction_time))
# get mean RTs in each condition for each subject
rt_data_subject_compatible <- all_sim_data %>%
mutate(odd_even = case_when(stimulus %% 2 == 0 ~ "even",
stimulus %% 2 != 0 ~ "odd"),
correct_response = case_when(odd_even == "odd" ~ "left",
odd_even == "even" ~ "right")) %>%
mutate(compatible = case_when(stimulus < 5 & correct_response == "left" ~ "compatible",
stimulus < 5 & correct_response == "right" ~ "incompatible",
stimulus > 5 & correct_response == "left" ~ "incompatible",
stimulus > 5 & correct_response == "right" ~ "compatible")
) %>%
mutate(accuracy = case_when(correct_response == response ~ TRUE,
correct_response != response ~ FALSE
)
) %>%
filter(accuracy == TRUE) %>%
group_by(compatible,sim_subject) %>%
summarize(mean_rt = mean(reaction_time), .groups = "drop")
# Compute difference scores for each subject
rt_data_subject_SNARC <- rt_data_subject_compatible %>%
pivot_wider(names_from = compatible,
values_from = mean_rt) %>%
mutate(SNARC_effect = incompatible-compatible)
# make plots
F1A <- ggplot(rt_data_subject_compatible, aes(x = compatible,
y = mean_rt))+
geom_violin()+
stat_summary(fun = "mean",
geom = "crossbar",
color = "red")+
geom_point()+
theme_classic(base_size=15)+
ylab("Mean Simulated Reaction Time") +
ggtitle("A")
F1B <- ggplot(rt_data_subject_SNARC, aes(x = ' ',
y = SNARC_effect))+
geom_violin()+
stat_summary(fun = "mean",
geom = "crossbar",
color = "red")+
geom_point()+
theme_classic(base_size=15)+
ylab("Simulated SNARC Effects")+
xlab("Incompatible - Compatible")+
ggtitle("B")
F1A + F1B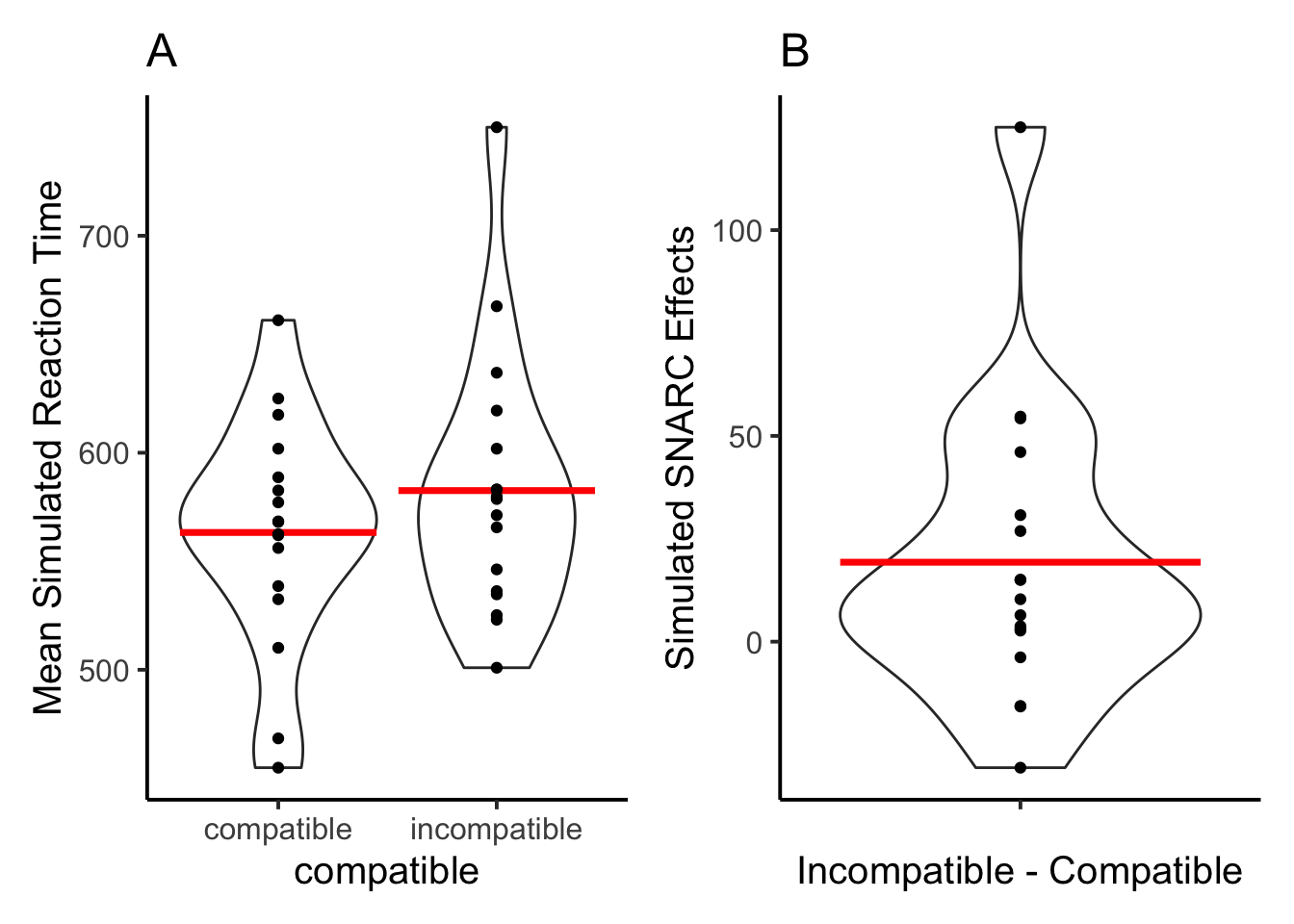
13 out of 17 simulated subjects showed a positive SNARC effect.
Here is a histogram of the individual simulated reaction times.

The prompt did not specify to produce values with different endings. As with previous simulations, the model prefers values ending in 0.
all_sim_data <- all_sim_data %>%
mutate(ending_digit = stringr::str_extract(all_sim_data$reaction_time, "\\d$")) %>%
mutate(ending_digit = as.numeric(ending_digit))
ggplot(all_sim_data, aes(x=ending_digit))+
geom_histogram(binwidth=1, color="white")+
scale_x_continuous(breaks=seq(0,9,1))+
theme_classic(base_size = 10)+
xlab("Simulated RT Ones Digit")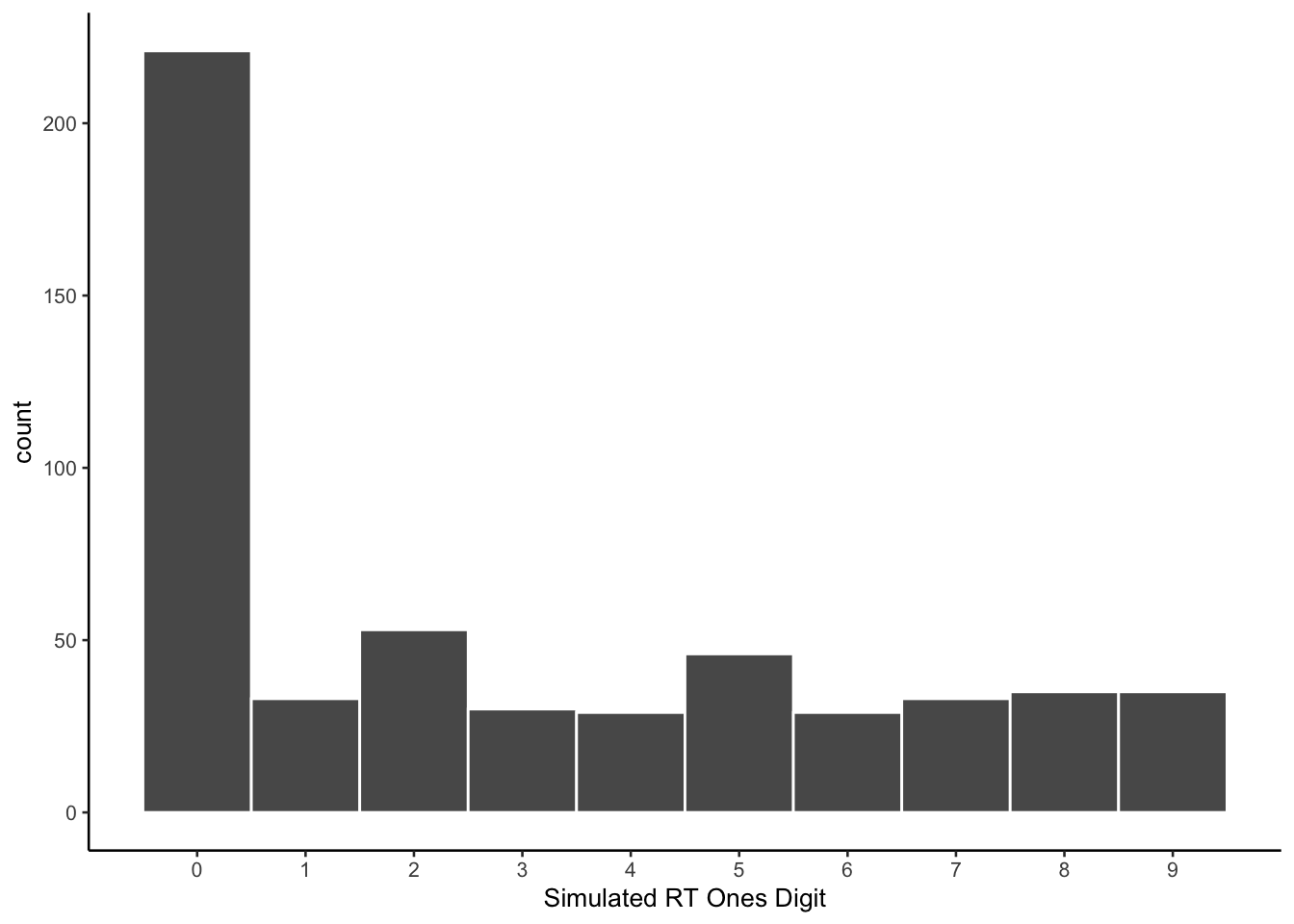
# report accuracy data
accuracy_data_subject <- all_sim_data %>%
mutate(odd_even = case_when(stimulus %% 2 == 0 ~ "even",
stimulus %% 2 != 0 ~ "odd"),
correct_response = case_when(odd_even == "odd" ~ "left",
odd_even == "even" ~ "right")) %>%
mutate(compatible = case_when(stimulus < 5 & correct_response == "left" ~ "compatible",
stimulus < 5 & correct_response == "right" ~ "incompatible",
stimulus > 5 & correct_response == "left" ~ "incompatible",
stimulus > 5 & correct_response == "right" ~ "compatible")
) %>%
mutate(accuracy = case_when(correct_response == response ~ TRUE,
correct_response != response ~ FALSE
)) %>%
group_by(compatible,sim_subject) %>%
summarize(proportion_correct = mean(accuracy), .groups = "drop")
ggplot(accuracy_data_subject, aes(x = compatible,
y = proportion_correct))+
geom_violin()+
stat_summary(fun = "mean",
geom = "crossbar",
color = "red")+
geom_point()+
theme_classic(base_size=15)+
ylab("Simulated Proportion Correct")+
xlab("Compatibility")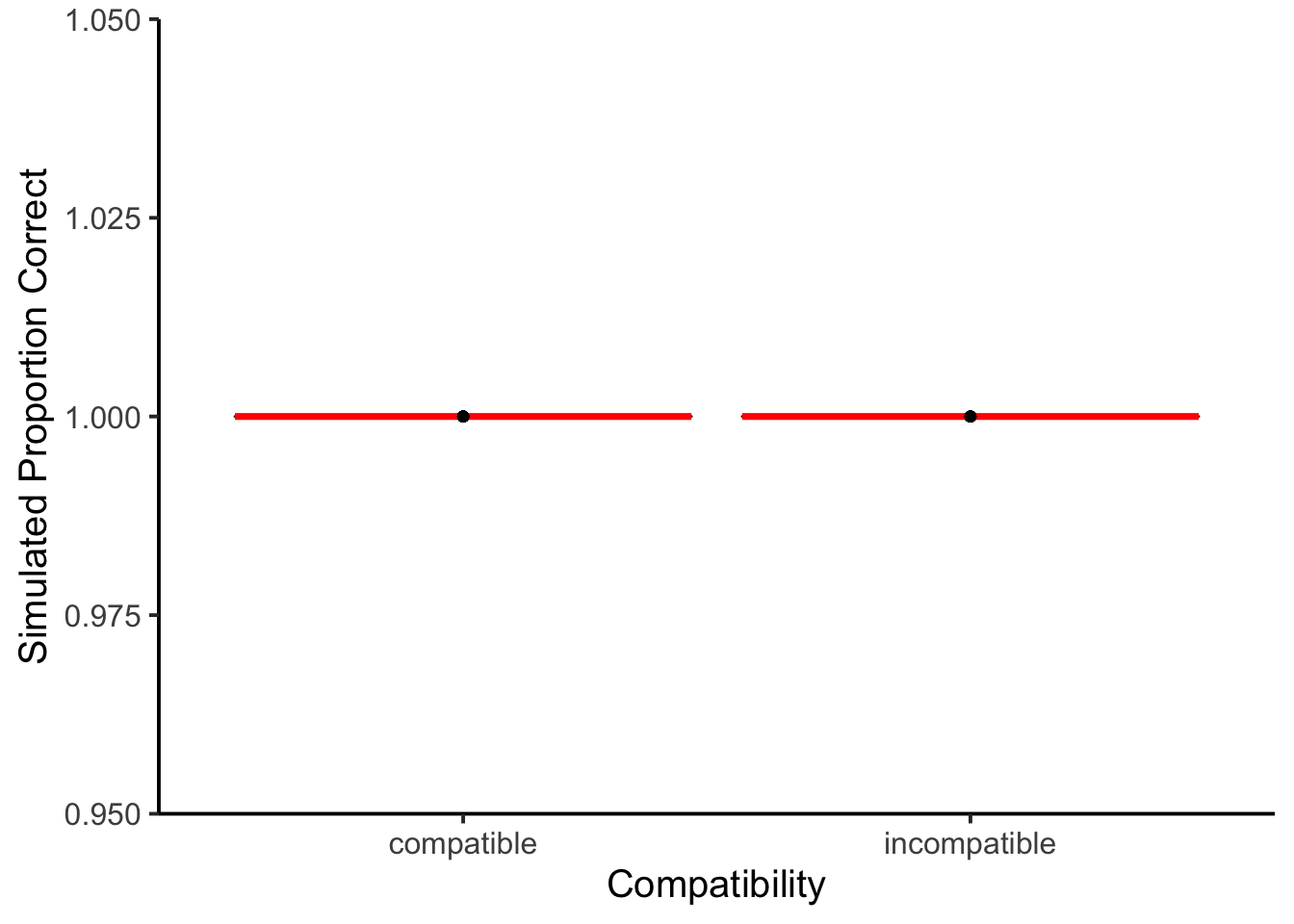
The model was always made correct responses.
It appears that the model produced a typical SNARC effect with minimal prompting. I did not counterbalance whether the odd or even response was left or right.